PLOS ONE: Absolute Quantitation of Met Using Mass Spectrometry for Clinical Application: Assay Precision, Stability, and Correlation with MET Gene Amplification in FFPE Tumor Tissue
![TMT Labeling for the Masses: A Robust and Cost-efficient, In-solution Labeling Approach*[S] - Molecular & Cellular Proteomics TMT Labeling for the Masses: A Robust and Cost-efficient, In-solution Labeling Approach*[S] - Molecular & Cellular Proteomics](https://els-jbs-prod-cdn.jbs.elsevierhealth.com/cms/attachment/e1426d50-a4d4-41ad-b5cb-f6710a6f63da/fx1_lrg.jpg)
TMT Labeling for the Masses: A Robust and Cost-efficient, In-solution Labeling Approach*[S] - Molecular & Cellular Proteomics

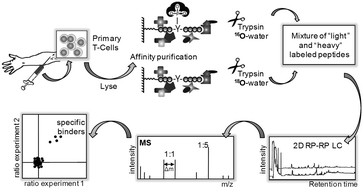
Illustration of stable isotope labeling and quantitative MS.Cells are... | Download Scientific Diagram

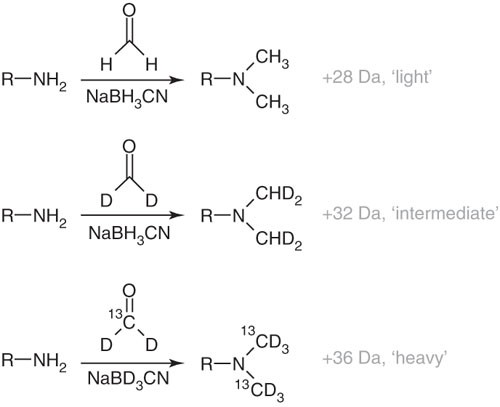
Stable isotope dimethyl labelling for quantitative proteomics and beyond | Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences
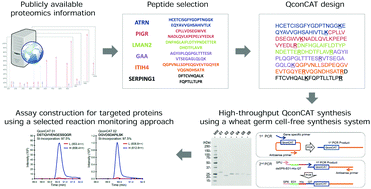
High-throughput production of a stable isotope-labeled peptide library for targeted proteomics using a wheat germ cell-free synthesis system - Molecular BioSystems (RSC Publishing)

Schematic work flow of quantitation by SRM coupled with stable isotope... | Download Scientific Diagram

Isotopically Labeled Clickable Glutathione to Quantify Protein S‐Glutathionylation - VanHecke - 2020 - ChemBioChem - Wiley Online Library

Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics | Nature Protocols

Mass spectrometry-based absolute quantification reveals rhythmic variation of mouse circadian clock proteins | PNAS